Project Description
Author: Ancheng Deng (myself)
Outstanding Undergraduate Thesis, 2017
School of Mathematics, SYSU
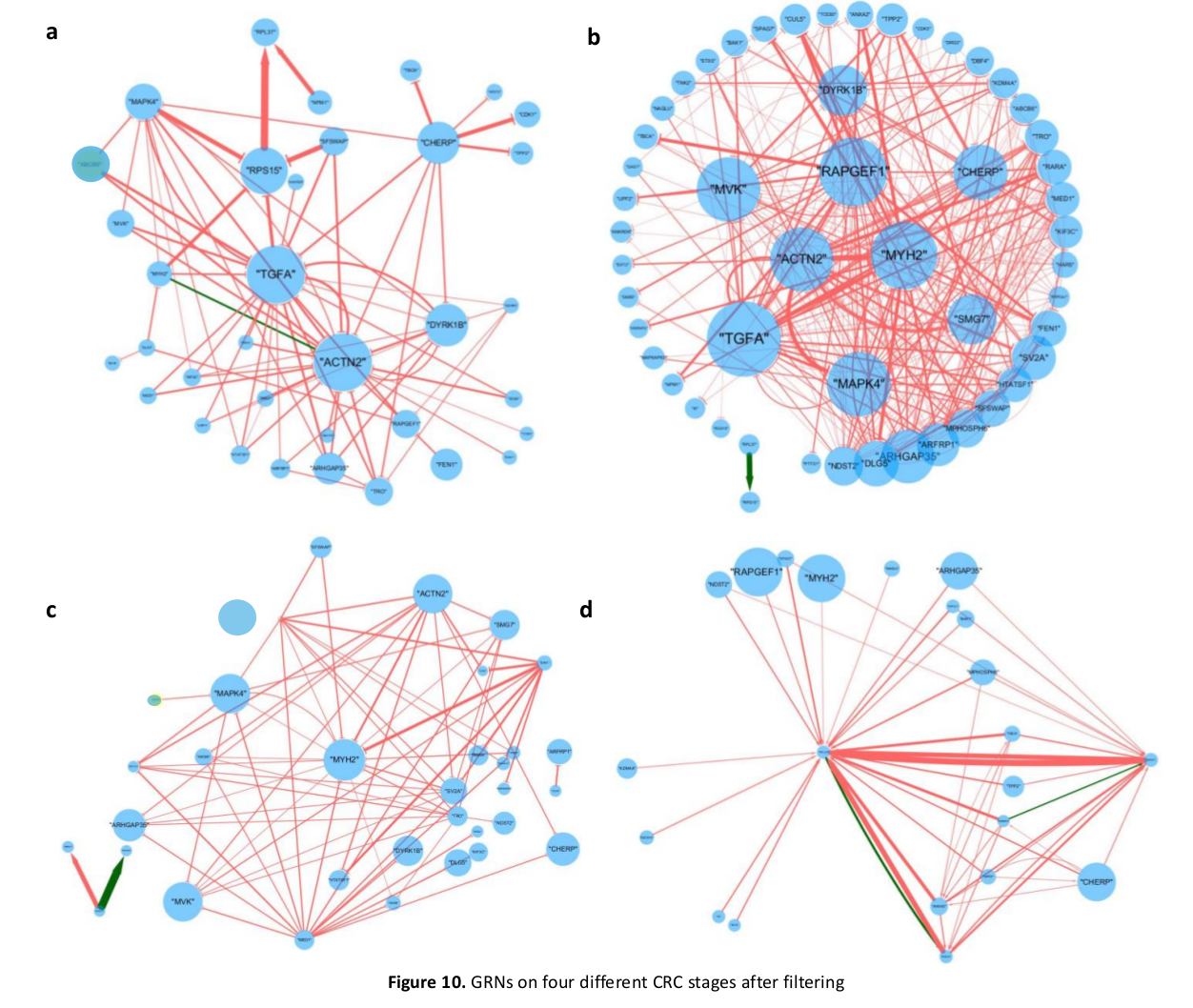
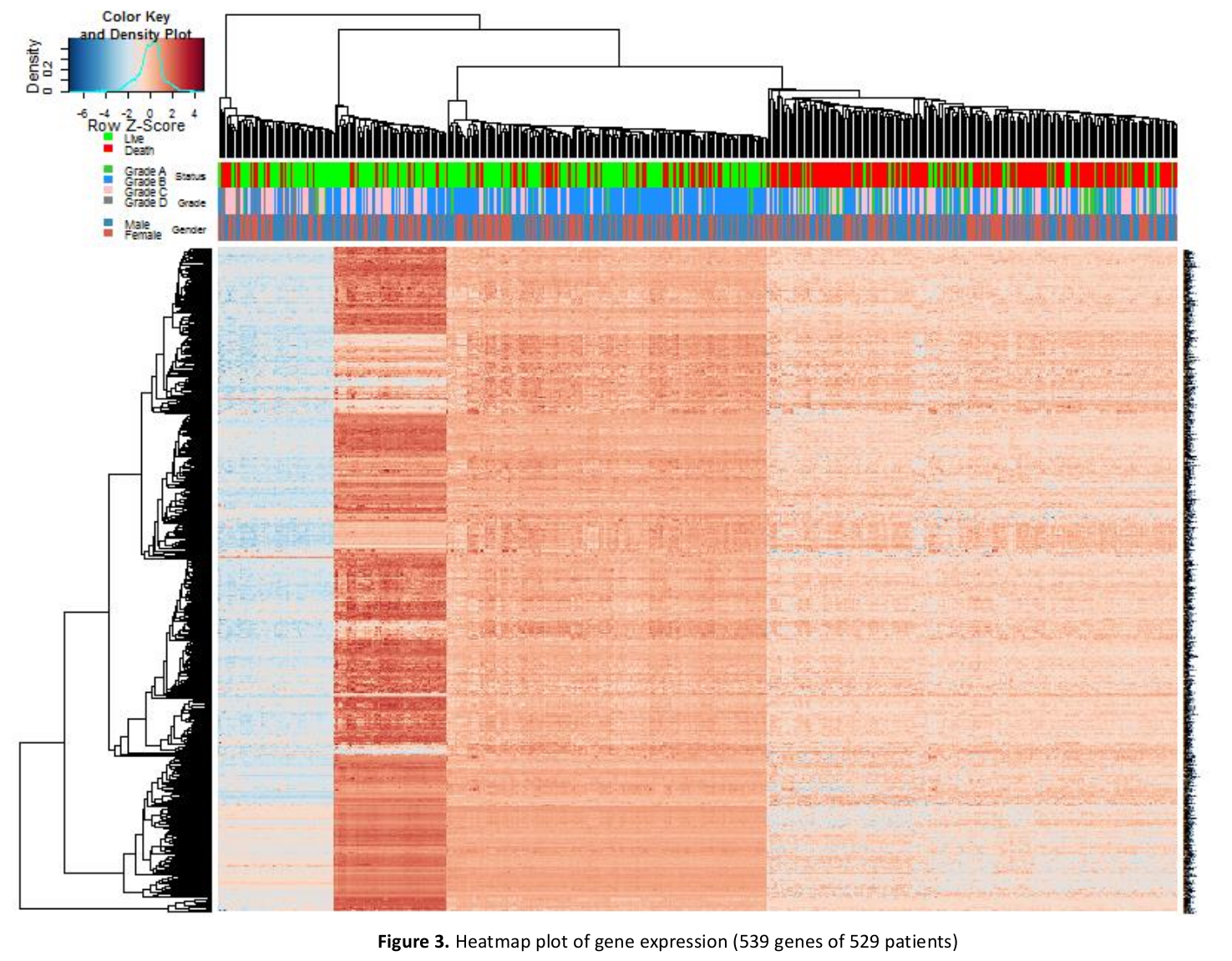
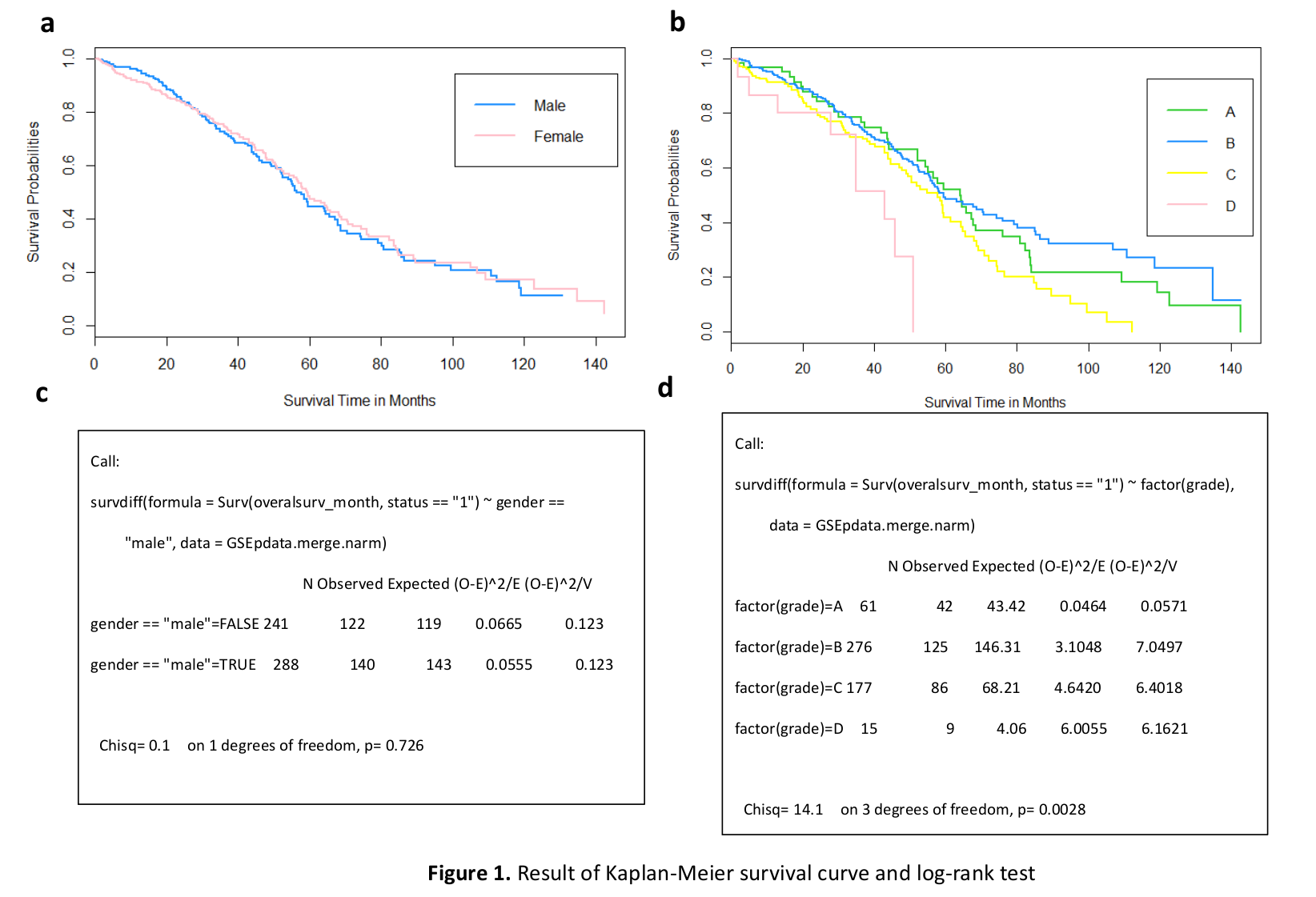
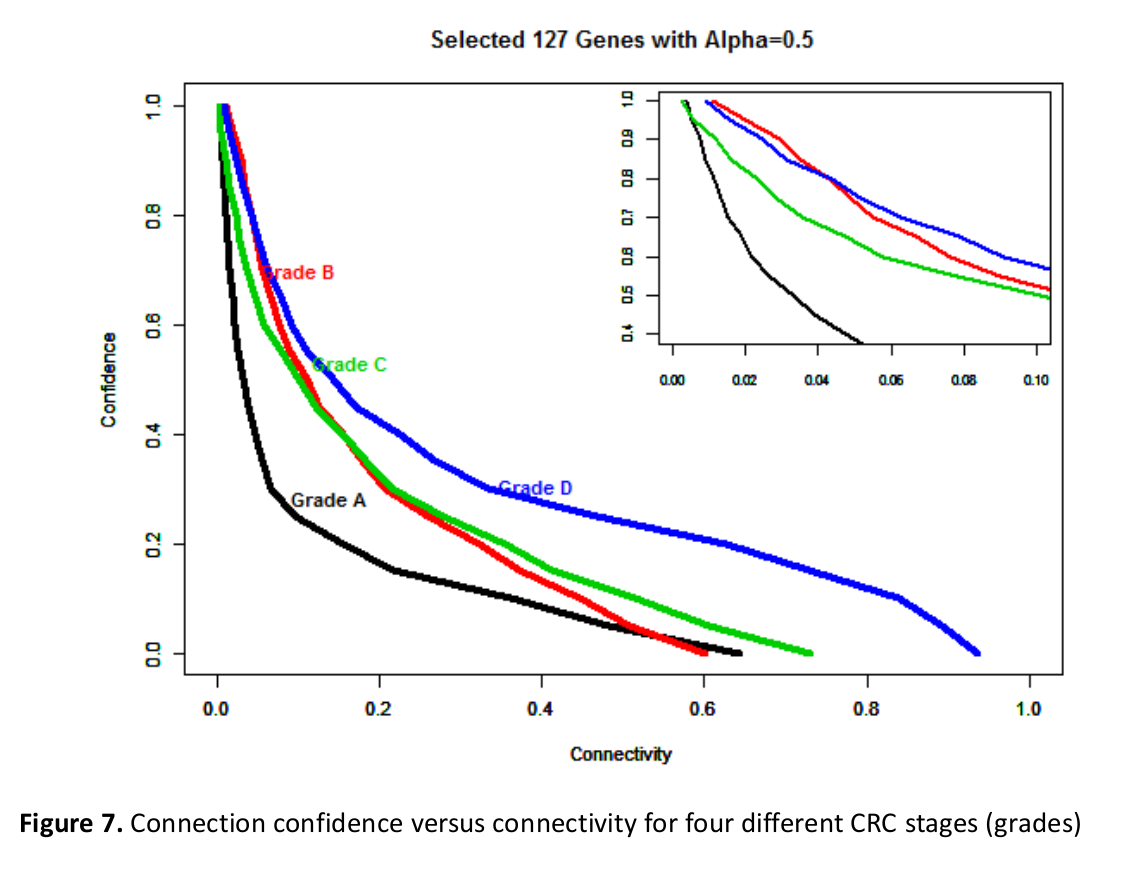
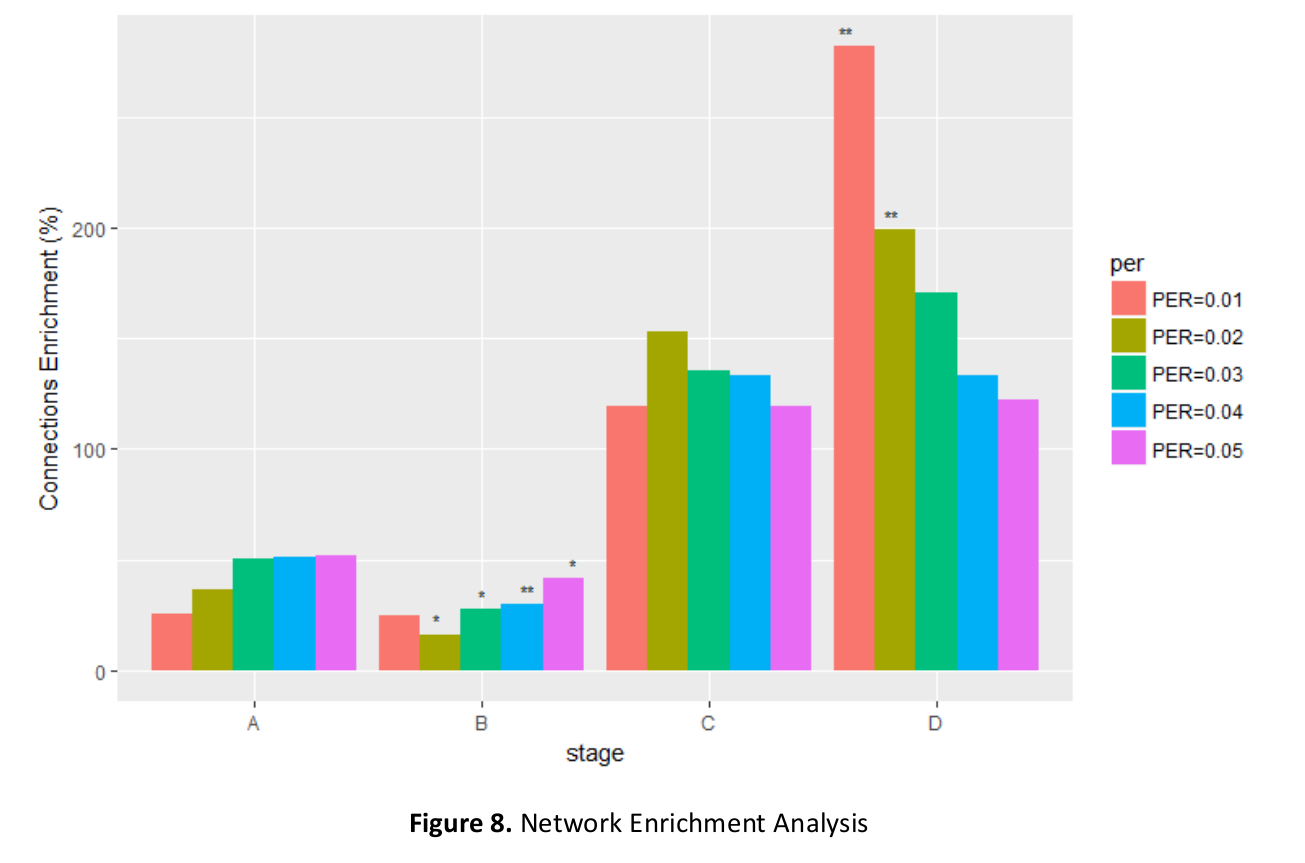
Abstract: In this paper we are attempting to reconstruct the dynamic gene regulation network from a combined colorectal dataset using one method called DCM (dynamic cascaded method). Mutual information network is utilized to have a broad view of the correlation of expression value, and a filter is added to further diminish number of potential important genes based on the sum of mutual information score. Finally, a set of 127 genes is obtained, and the DCM method is implemented on our 127 genes set to reconstruct the dynamic GRN (gene regulatory network). Multiple attributes can be contained in this network, such as up/down regulation, strength of the regulation, confidence of a connection and the degree of nodes (genes). Reconstructing dynamic GRNs based on different colorectal cancer stages allows us to follow the dynamic trend of gene regulation as well as find novel target genes for medicine development and gene therapy.
Data Source: The combined dataset is obtained from 4 sepearate sources: Vanderbilt University (GSE17536), Ludwig Institute for Cancer Research (GSE14333), Signature Diagnostics AG (GSE12945) and Chinese hospital (patient67).
Gene Regulatory Network Analysis Report